Virtual Screening
Unlocking the Future of Drug Discovery: Precision Virtual Screening for Targeted Therapeutics
Accelerating Drug Development via Virtual Screening
Virtual screening is a computational method used to predict how small molecules, like drugs, interact with target proteins. It involves simulating molecular docking to identify potential drug candidates from large chemical libraries, reducing the need for physical testing.
Virtual screening is used in drug discovery to identify promising compounds, assist in drug repurposing, and develop personalized treatments. It speeds up the process of finding effective and safe drugs by predicting interactions before lab experiments.
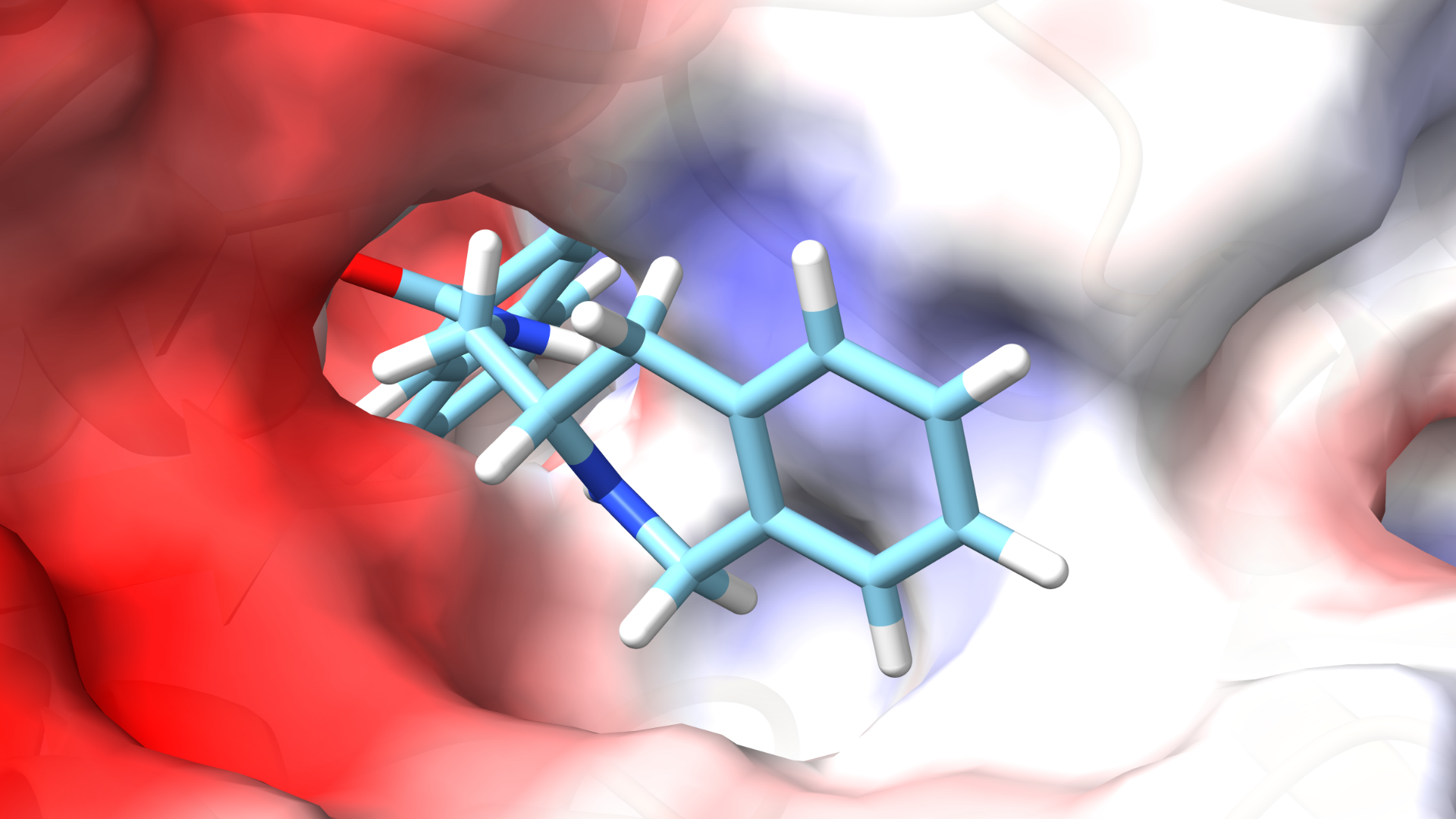
Virtual Screening of Natural Products: A Path to Novel Treatments for Breast and Lung Cancer Mutations
Lung cancer is the leading cause of cancer-related deaths among men worldwide, while breast cancer holds the same position among women. Understanding the molecular biology of these cancers is crucial, particularly by identifying and analyzing the somatic mutations that drive their progression.
Our research focuses on virtual screening of natural product compound libraries to identify potential inhibitors targeting proteins encoded by the top 20 mutated genes in breast and lung cancers. Natural products offer a diverse array of bioactive molecules with unique chemical structures, making them invaluable in drug discovery. By docking these compounds onto selected proteins associated with critical mutations, we aim to uncover new inhibitors that can modulate aberrant protein functions, potentially leading to effective therapies for these prevalent cancers.
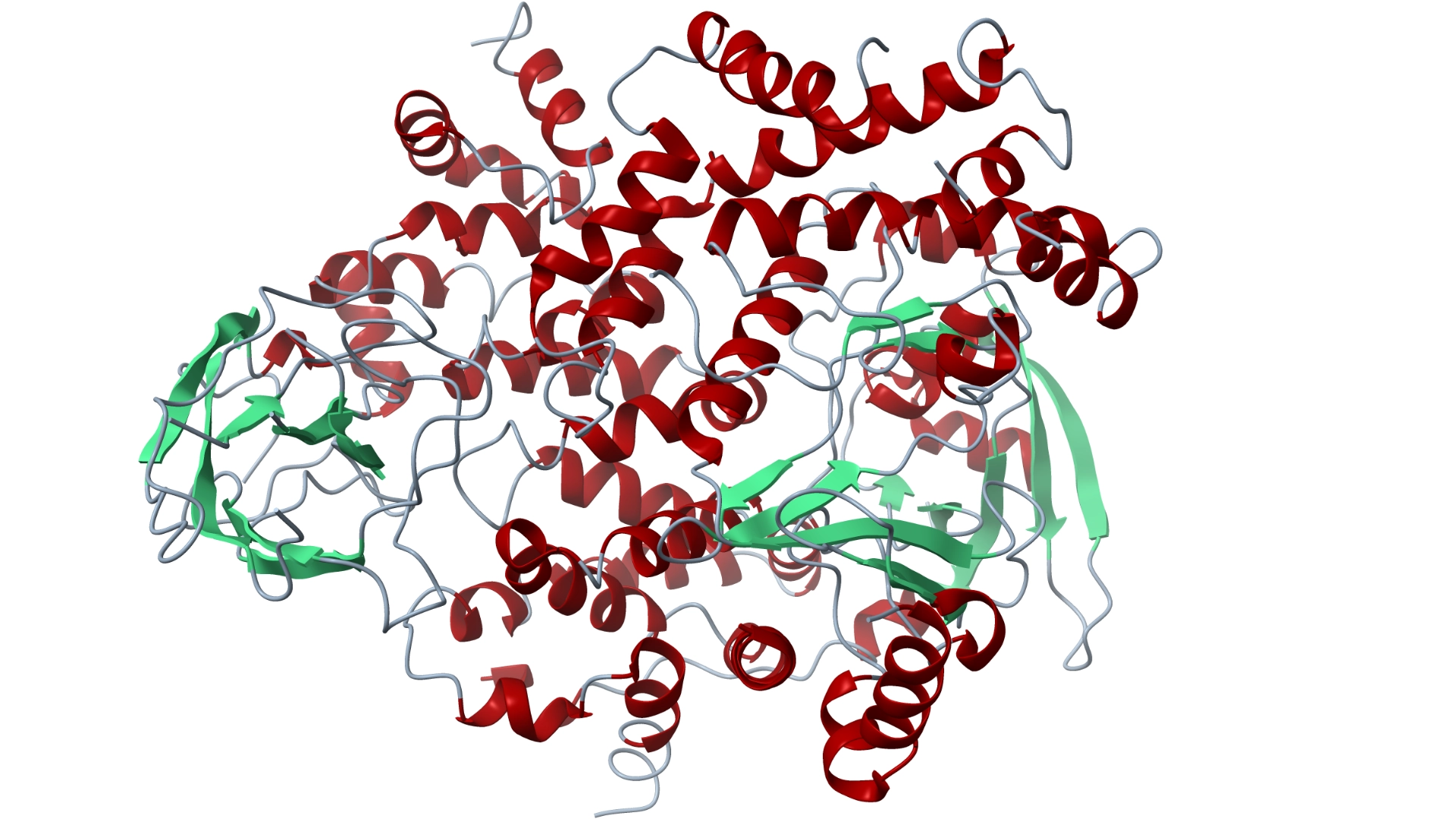
Targeting Aldehyde Dehydrogenase: A Virtual Screening Approach Using Natural Product Databases
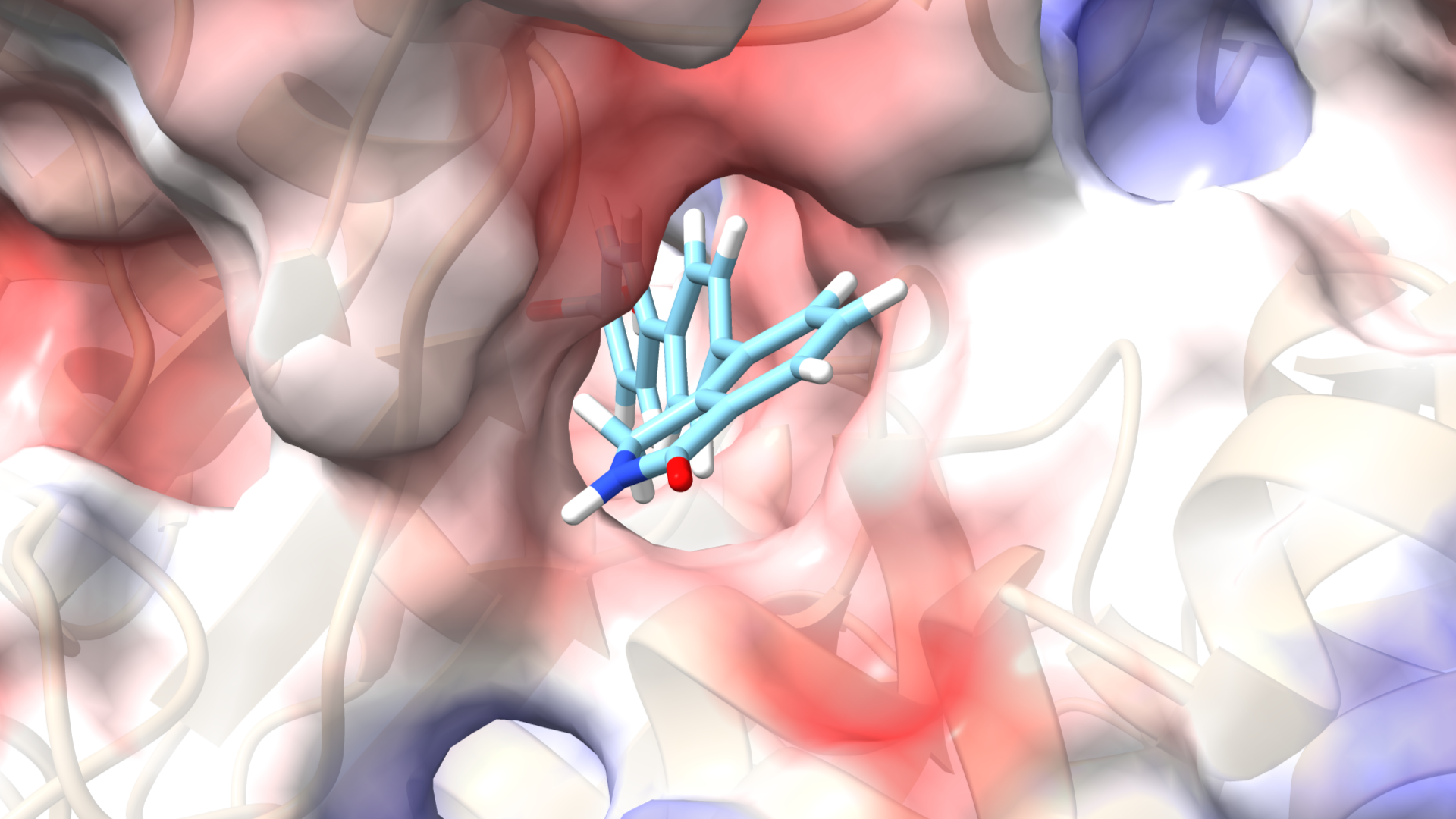
Aldehyde dehydrogenases (ALDHs) are a family of enzymes that catalyze the oxidation of aldehydes to carboxylic acids, playing a vital role in detoxification and metabolism. Found in various tissues, ALDHs help process toxic byproducts, including those from alcohol metabolism. Alterations in specific ALDH isoforms are linked to diseases such as cancer, neurodegenerative disorders, and metabolic syndromes.
Inhibiting ALDH proteins offers significant therapeutic benefits, particularly in cancer and neuroprotection. Some ALDH isoforms are linked to cancer stem cells, contributing to tumor growth and treatment resistance. Targeting these enzymes can enhance cancer therapy efficacy. Additionally, inhibiting specific ALDHs may reduce oxidative stress, making it a potential strategy for neurodegenerative diseases. Overall, ALDH inhibition can provide insights into metabolic pathways and address conditions involving aldehyde accumulation.
Our research group is conducting a virtual screening study using natural product databases to identify potential ALDH inhibitors. Utilizing AutoDock Vina, we will simulate docking interactions between these natural compounds and various ALDH isoforms to predict binding affinities. This in silico approach aims to discover novel inhibitors for therapeutic applications in cancer treatment and other ALDH-related conditions, leveraging the diversity of natural products.
Molecular Docking of Urease Inhibitors: A Step Towards New Treatments for Gastric Ulcers
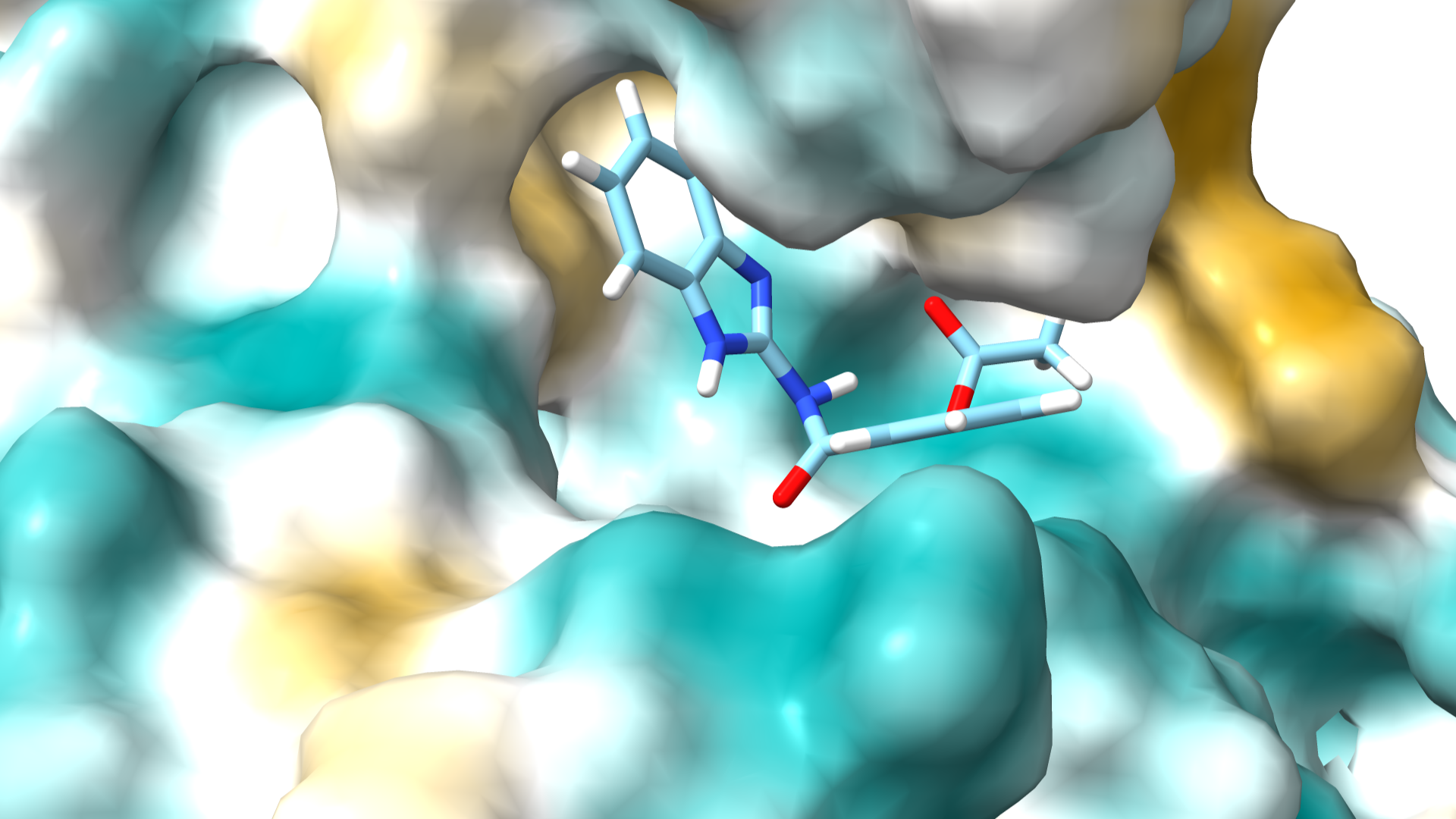
Urease is a metalloenzyme that catalyzes the hydrolysis of urea into ammonia and carbon dioxide, playing a vital role in nitrogen metabolism. Produced by pathogenic bacteria like Helicobacter pylori, urease raises local pH levels, facilitating bacterial survival in the acidic stomach. Understanding its structure and function is crucial for developing targeted inhibitors to combat its pathogenic effects.
Inhibiting urease offers significant therapeutic benefits, particularly for gastrointestinal health. Reduced urease activity lowers ammonia production, decreasing local pH and enhancing the efficacy of antibiotics against Helicobacter pylori. This leads to less gastric inflammation and ulcer formation. Additionally, urease inhibitors can improve nitrogen use in agriculture, reducing environmental impact. Thus, effective urease inhibitors are promising for both medical and agricultural applications.
Our research group is conducting a study using AutoDock Vina to identify potential inhibitors from a local library of compounds. This in silico approach predicts binding affinities and interactions between compounds and urease, guiding the development of novel therapeutics aimed at improving treatment outcomes for gastric ulcers and related disorders.
