Molecular Dynamics Simulations
Simulating Molecular Interactions to Uncover Biological Insights
Molecular Dynamics Simulations: Advancing Science, Medicine, and Drug Discovery
Molecular dynamics (MD) simulations are computational techniques used to study the behavior of atoms and molecules over time. They model the physical movements and interactions of particles according to classical physics principles, such as Newton’s laws of motion. By simulating the interactions between particles based on classical mechanics, these simulations provide insights into the behavior of complex systems at the atomic and molecular levels.
MD simulations are a powerful tool across various fields. In science, they deepen our understanding of material properties and biological processes. In medicine and pharmaceuticals, they are crucial for elucidating disease mechanisms, designing and optimizing drugs, and advancing personalized medicine. By providing detailed atomic-level insights, MD simulations enable researchers to make informed decisions and accelerate discovery and development processes.
Advancing Cancer Research through Gaussian Accelerated Molecular Dynamics
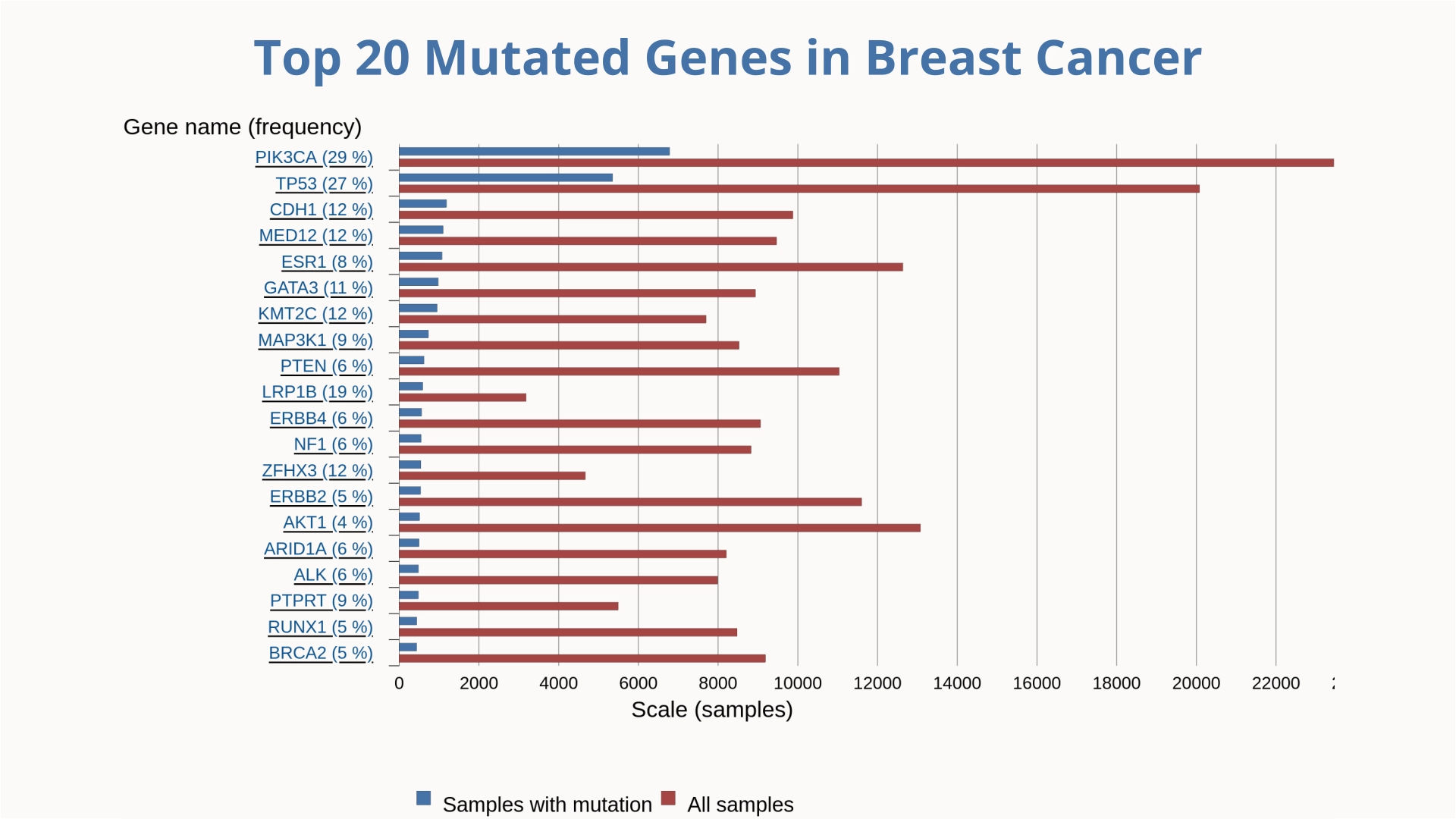
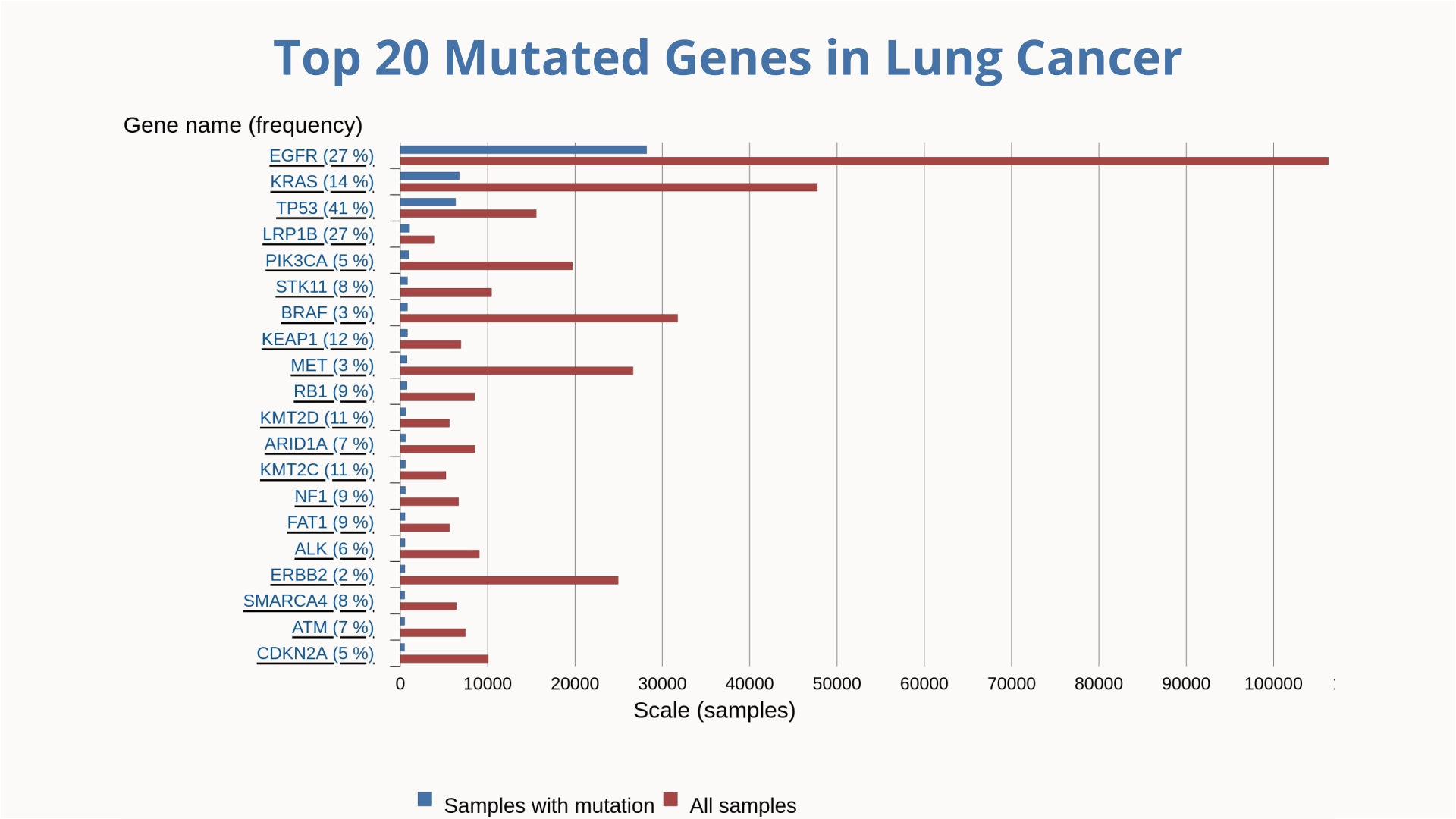
Lung cancer remains the leading cause of cancer-related deaths among men globally, while breast cancer holds the same position among women. Understanding the molecular biology of cancer requires identifying and analyzing the mutations that drive these diseases.
Gaussian Accelerated Molecular Dynamics (GaMD) is a powerful computational technique that enhances the sampling efficiency of molecular dynamics simulations, enabling the exploration of complex biomolecular processes.
In our research, we employ GaMD to investigate the dynamic behavior of targeted proteins among the top 20 mutant genes implicated in breast and lung cancers. This approach allows us to capture rare conformational changes and transient states of these proteins, providing deeper insights into their functional mechanisms and potential roles in oncogenesis. Additionally, we utilize GaMD to study protein-ligand interactions, facilitating the identification of novel therapeutic compounds with improved binding affinities and specificity.
